우리 학과 송지준 교수와 제1저자 조수민 연구교수, 장주원 박사과정 학생이 nature communications에 “Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.”로 논문이 게재되었습니다.

Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Abstract
The fundamental unit of chromatin, the nucleosome, is an intricate structure that requires histone chaperones for assembly. ATAD2 AAA+ ATPases are a family of histone chaperones that regulate nucleosome density and chromatin dynamics. Here, we demonstrate that the fission yeast ATAD2 homolog, Abo1, deposits histone H3-H4 onto DNA in an ATP-hydrolysis-dependent manner by in vitro reconstitution and single-tethered DNA curtain assays. We present cryo-EM structures of an ATAD2 family ATPase to atomic resolution in three different nucleotide states, revealing unique structural features required for histone loading on DNA, and directly visualize the transitions of Abo1 from an asymmetric spiral (ATP-state) to a symmetric ring (ADP- and apo-states) using high-speed atomic force microscopy (HS-AFM). Furthermore, we find that the acidic pore of ATP-Abo1 binds a peptide substrate which is suggestive of a histone tail. Based on these results, we propose a model whereby Abo1 facilitates H3-H4 loading by utilizing ATP.
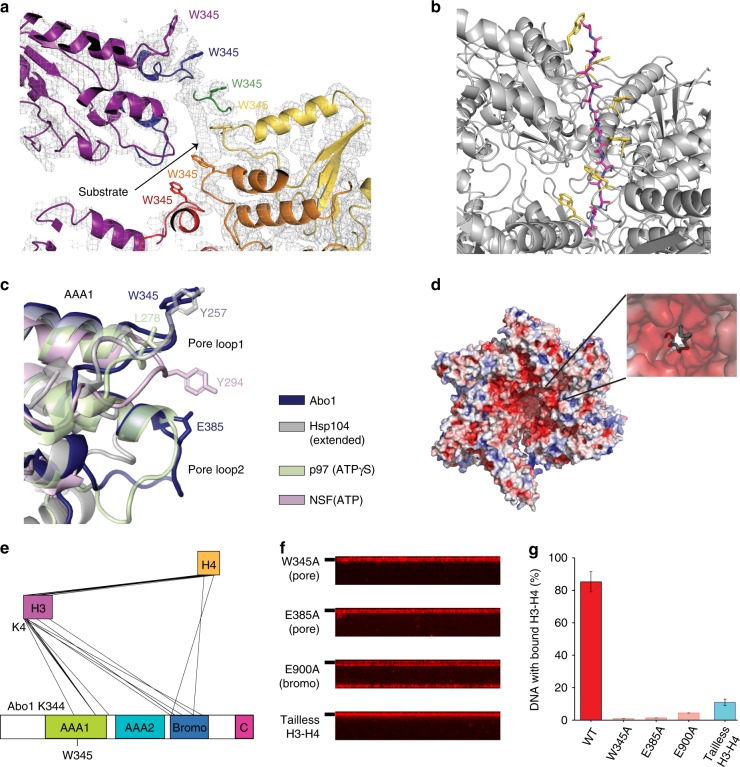
a Extra density near the Abo1 central pore region, surrounded by Abo1 W345 tryptophan side chains. b Polyalanine model of the substrate peptide (magenta) built into the extra electron density observed in a, with W345 side chains colored in yellow. c Superposition of Abo1 AAA1 pore loops 1 & 2 with other AAA+ ATPases. Abo1 (dark blue), Hsp104 in the extended state, (gray, PDB ID: 5VYA), p97 in the ATPγS state (light green, PDB ID: 5FTL), and NSF in the ATP state (pink, PDB ID: 3J94) were aligned by AAA1 NBD. Key substrate binding residues on pore loop 1 and the acidic residue of pore loop2, E385, are labeled. d Electrostatic surface representation of Abo1 highlighting the negatively charged pore interior contributed by acidic pore loop residues. e Depiction of intermolecular crosslinks between Abo1 and histone H3–H4 detected by XL-MS and filtered with a crosslink confidence (LD) score cutoff of 20. The highest LD score intermolecular crosslink, a crosslink between H3 the N-terminus (K4) and Abo1 AAA1 pore loop1 (K344), is labeled. f DNA curtain-based H3–H4 deposition assays performed with Abo1 pore loop mutants (W345A and E385A) and a bromodomain mutant (E900A) with wild-type H3–H4, and H3–H4 deposition assays of wild-type Abo1 with N-terminal tail truncated H3–H4. g Comparison of H3–H4 loading activity on DNA by quantification of fraction DNA bound with labeled H3–H4. Error bars represent SD from three experiments. The numbers of molecules analyzed for each condition are n = 244 for WT, n = 222 for W345A, n = 219 for E385, n = 249 for E900A and n = 238 for tailless H3–H4.
https://www.ncbi.nlm.nih.gov/pubmed/31848341








